BIGDATA: Analytical Approaches to Massive Data Analysis
Department of Computer Science — Brown University
Project Description
The goal of this project is to design and test mathematically well-founded algorithmic and statistical techniques for analyzing large scale, heterogeneous and noisy data. The proposed research is transformative in its emphasis on rigorous analytical evaluation of algorithms' performance and statistical measures of output uncertainty, in contrast to the primarily heuristic approaches currently used in data mining and machine learning. Any progress in that direction will have significant contribution to the reliability and scientific impact of massive data analysis. This project is motivated by the challenges in analyzing molecular biology data. Molecular biology provides an excellent source of data for testing advanced data analysis techniques: specifically, DNA/RNA sequence data repositories are growing at a super-exponential rate. The data is typically large and noisy, and in some cases includes both genotype and phenotype features that permits experimental validation of the analysis. However, the methods and techniques developed in this project will be broadly applicable to other scientific communities that process massive multi-variant data sets.
The major technical goals of the project include: (1) Design efficient algorithms that provide guarantees on the output when the data comes from independent random samples from an unknown distribution. (2) Develop techniques for estimating the minimum number of samples required to test hypothesis of varying complexity in large datasets, building on techniques in computational statistics. (3) Design algorithms to analyze data on graphs that represent interactions between samples or features in the dataset. These data may be static (e.g. mutations on interacting genes represented by a protein interaction network) or dynamic (e.g. information dissemination on a social network).
This project will advocate a responsible approach to data analysis, based on well-founded mathematical and statistical concepts. The capacity building activities of the project include: (1) Creation and dissemination of algorithms and software that implement rigorous computational and statistical approaches to big data analysis. (2) Educational initiatives at the graduate and undergraduate level to build a bigger workforce of data scientists with the appropriate foundational skills both to apply analytical tools to existing datasets and to develop new approaches to future datasets. The proposed work will be tested on extensive cancer genome data, contributing to health IT, one of the National Priority Domain Areas.
Participants
Faculty

P.I. Eli Upfal
Ph.D. Students
Alumni
Shahrzad Haddadan, Cyrus Cousins, Lorenzo De Stefani, Megumi Ando, Benedetto Jacopo Buratti, Patrick Clay, Alessandro Epasto, Rajesh Jayaram, Vidur Joshi, Evgenios M. Kornaropoulos, Ahmad Mahmoody, Sung-Ho (Justin) Oh, Maithra Raghu, Matteo Riondato, Henry Wallace
Publications
- A. Mazzetto, C. Menghini, A. Yuan, E. Upfal, and S. Bach. Tight Lower Bounds on Worst-Case Guarantees for Zero-Shot Learning with Attributes., to appear in NeurIPS 2022.
- C. Menghini, J. Uhr, S. Haddadan, A. Champagne, B. Sandstede, and S. Ramachandran. The Drift of# MyBodyMyChoice Discourse on Twitter, Runner-up for Best Paper, Websci 2022
- S. Haddadan, Y. Zhuang, C. Cousins, and E. Upfal Fast Doubly-Adaptive MCMC to Estimate the Gibbs Partition Function with Weak Mixing Time Bounds, NeurIPS 2021.
- L. De Stefani, E. Terolli, and E. Upfal. Tiered Sampling: An Efficient Method for Counting Sparse Motifs in Massive Graph Streams. ACM Trans. Knowl. Discov. Data 15, 2021
- C. Cousins, C. Wohlgemuth, M. Riondato. Bavarian: Betweenness Centrality Approximation with Variance-Aware Rademacher Averages, KDD 2021.
- A. Mazzetto*, C. Cousins*, D. Sam, S. Bach, E. Upfal. Adversarial Multi Class Learning under Weak Supervision with Performance Guarantees, ICML 2021.
- E. A. Viqueira, C. Cousins, A. Greenwald. Learning Competitive Equilibria in Noisy Combinatorial Markets, AAMAS 2021.
- S. Haddadan, C. Menghini, M. Riondato, E. Upfal. RePBubLik: Reducing Polarized Bubble Radius with Link Insertions, Runner-up for Best Paper, WDSM 2021.
- M. Ando, A. Lysyanskaya, and E. Upfal. On the Complexity of Anonymous Communications Through Public Networks, ITC 2021.
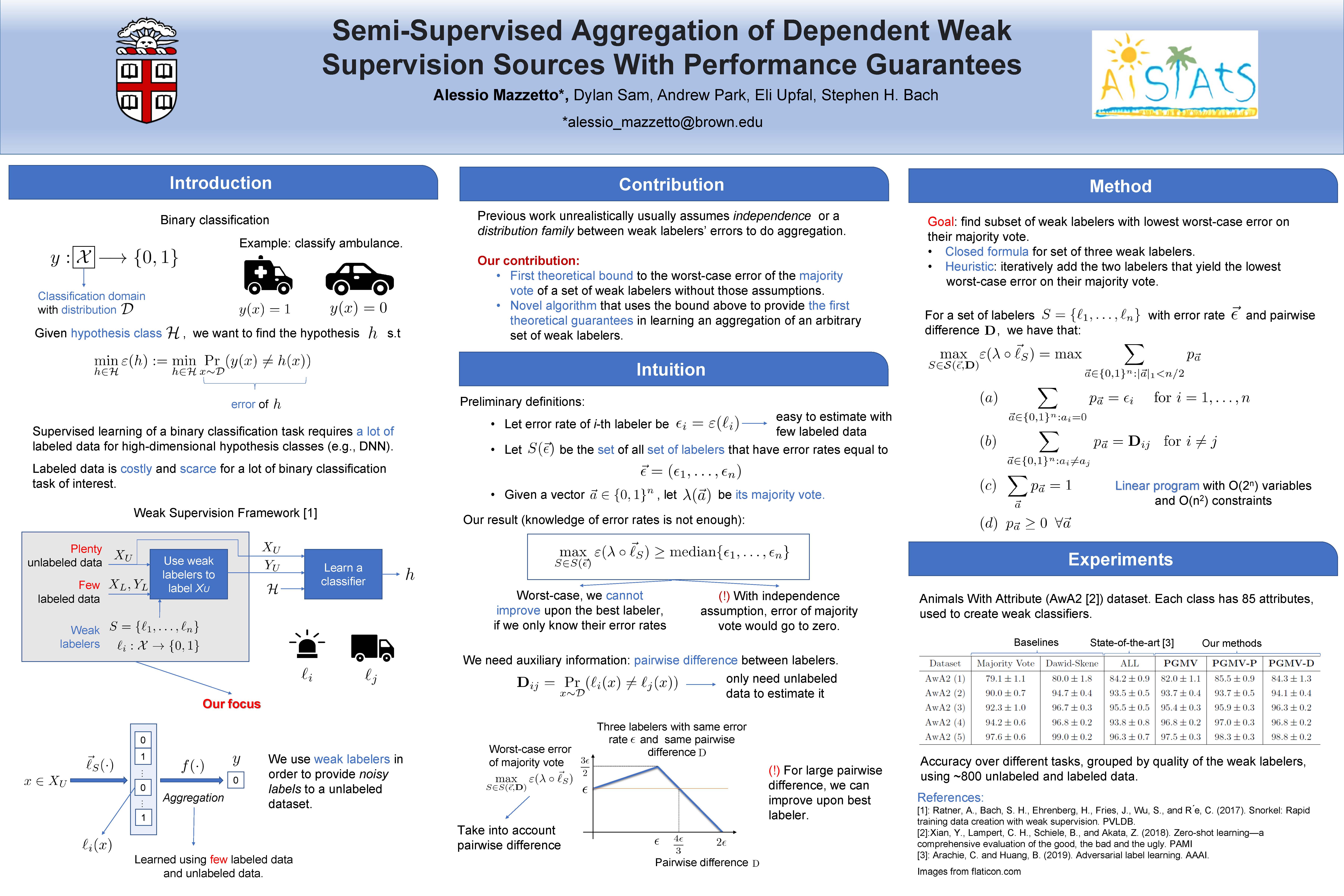
- A. Mazzetto, D. Sam, A. Park, E. Upfal, S. Bach. Semi-Supervised Aggregation of Dependent Weak Supervision Sources With Performance Guarantees, AISTATS 2021.
- L. Pellegrina, C. Cousins, F. Vandin, M. Riondato. MCRapper: Monte-Carlo Rademacher averages for poset families and approximate pattern mining, KDD 2020.
- E. A. Viqueira, C. Cousins, A. Greenwald. Improved Algorithms for Learning Equilibria in Simulation-Based Games, AAMAS 2020.
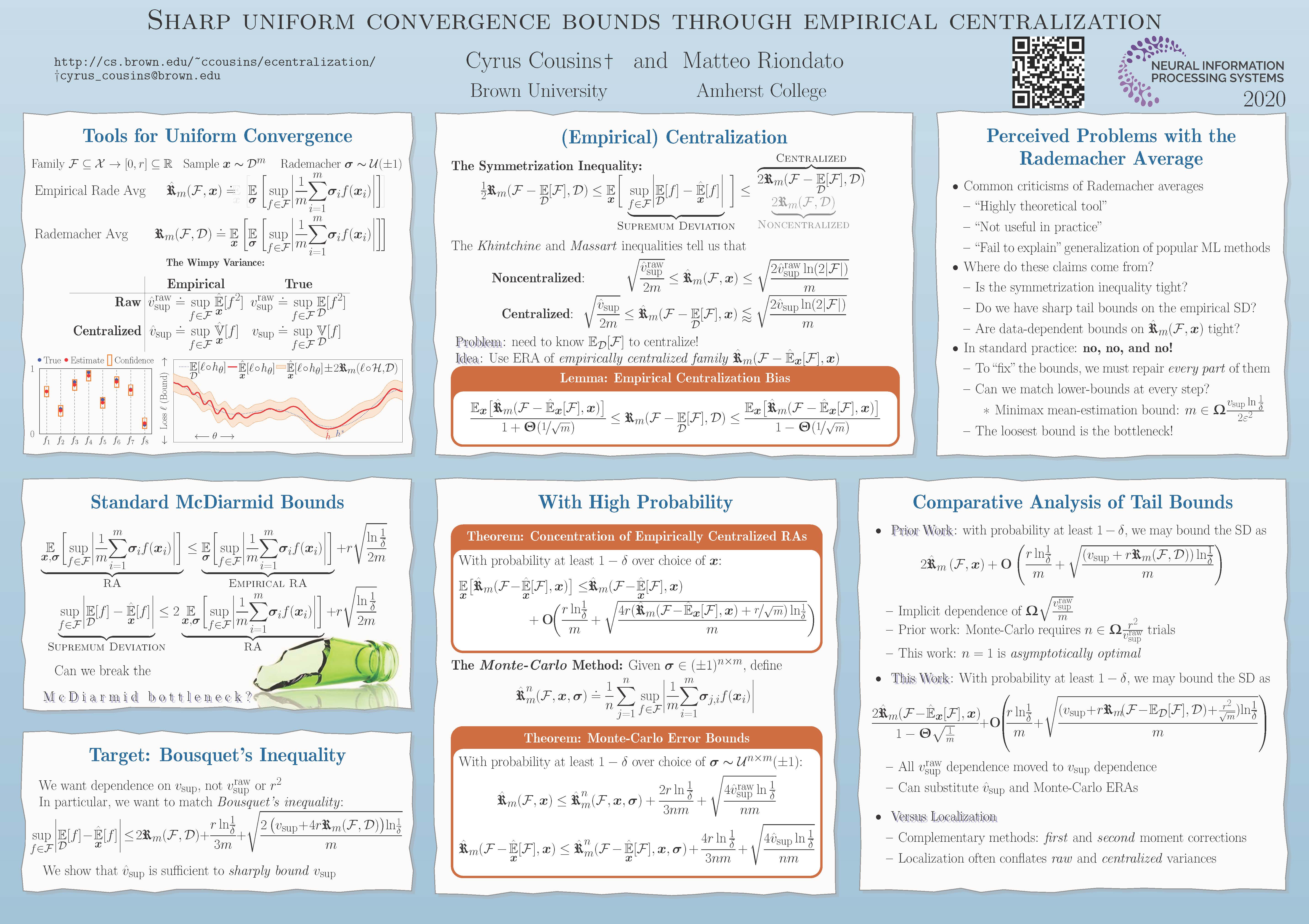
- C. Cousins, M. Riondato. Sharp uniform convergence bounds through empirical centralization, NeurIPS 2020.
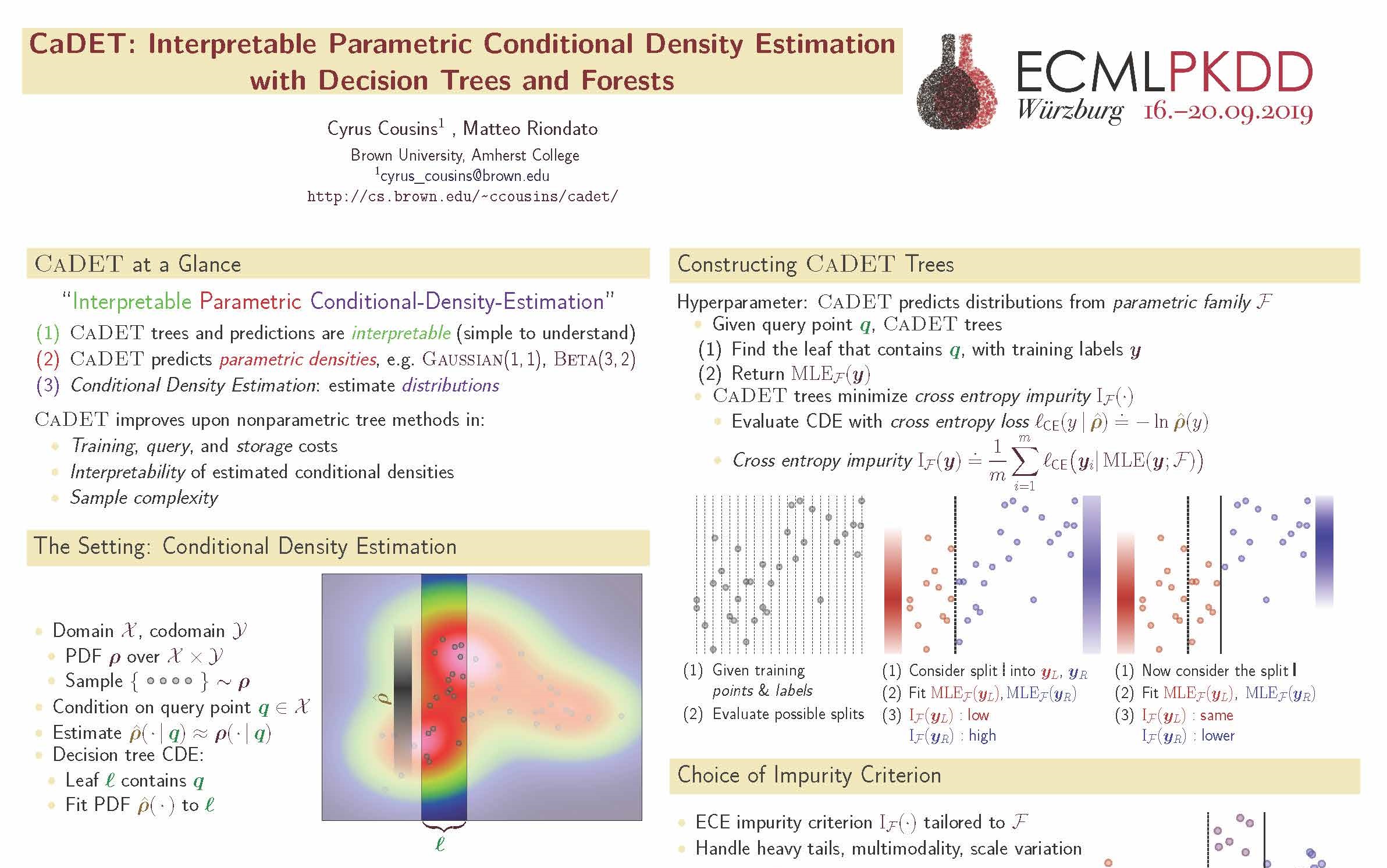
- C. Cousins, M. Riondato. CaDET: interpretable parametric conditional density estimation with decision trees and forests, ECML PKDD 2019.
- E. A. Viqueira, A. Greenwald, C. Cousins, E. Upfal. Learning Simulation-Based Games from Data, AAMAS 2019.
- E. A. Viqueira, C. Cousins, Y. Mohammad, A. Greenwald. Empirical Mechanism Design: Designing Mechanisms from Data. , UAI 2019.
- C. Menghini, A. Anagnostopoulos, E. Upfal, Wikipedia Polarization and Its Effects on Navigation Paths, IEEE BigData 2019.
- C. Cousins, E. Upfal. The k -Nearest Representatives Classifier: A Distance-Based Classifier with Strong Generalization Bounds. DSAA 2017
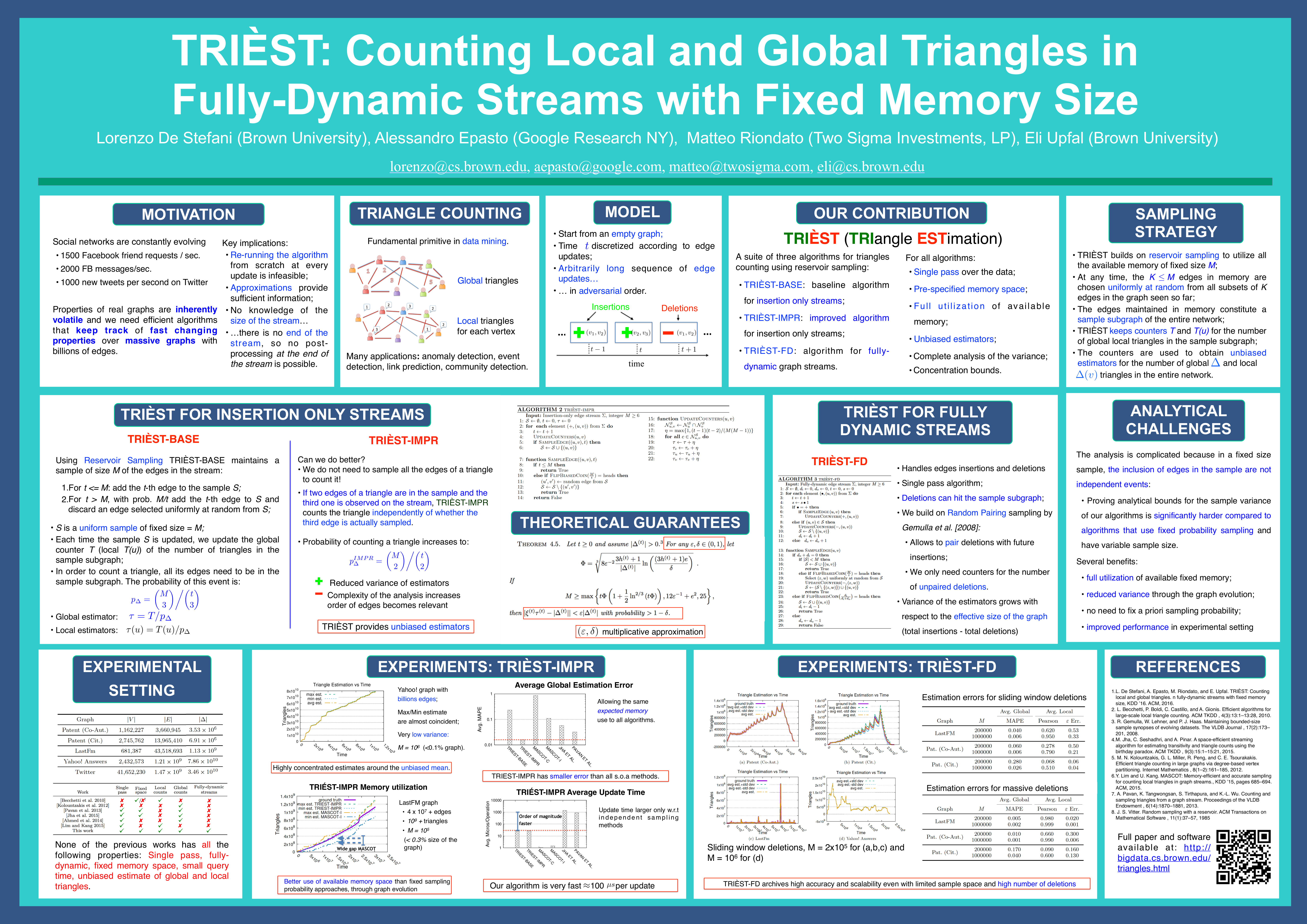
- L. De Stefani, A. Epasto, M. Riondato, E. Upfal. TRIÈST: Counting Local and Global Triangles in Fully Dynamic Streams with Fixed Memory Size. TKDD 11(4): 43:1-43:50 (2017)
- L. De Stefani, Z. Zhao, E. Zgraggen, C. Binnig, E. Upfal, T. Kraska. Controlling False Discoveries During Interactive Data Exploration. SIGMOD Conference 2017: 527-540
- Z. Zhao, E. Zgraggen, L. De Stefani, C. Binnig, E. Upfal, T. Kraska. Safe Visual Data Exploration, SIGMOD Conference 2017: 1671-1674
- M. Ceccarello, A. Pietracaprina, G. Pucci, E. Upfal. MapReduce and Streaming Algorithms for Diversity Maximization in Metric Spaces of Bounded Doubling Dimension, PVLDB 10(5): 469-480, 2017.
- C. Binnig, L. De Stefani, T. Kraska, E. Upfal, E. Zgraggen, Z. Zhao. Toward Sustainable Insights, or Why Polygamy is Bad for You, CIDR 2017.
- A. Epasto, S. Lattanzi, V. Mirrokni, I. Sebe and S. Verma. Ego-net Community Mining Applied to Friend Suggestion, VLDB 2016.
- M. Riondato, D. García-Soriano, F. Bonchi. Graph Summarization with Quality Guarantees, Data Mining and Knowledge Discovery, in press.
- L. De Stefani, A. Epasto, M. Riondato, E. Upfal. TRIÈST: Counting Local and Global Triangles in Fully-dynamic Streams with Fixed Memory Size, Best Student Paper Award, Research Track ACM KDD 2016.
- M. Riondato, E. Upfal. ABRA: Approximating Betweenness Centrality in Static and Dynamic Graphs with Rademacher Averages, ACM KDD 2016.
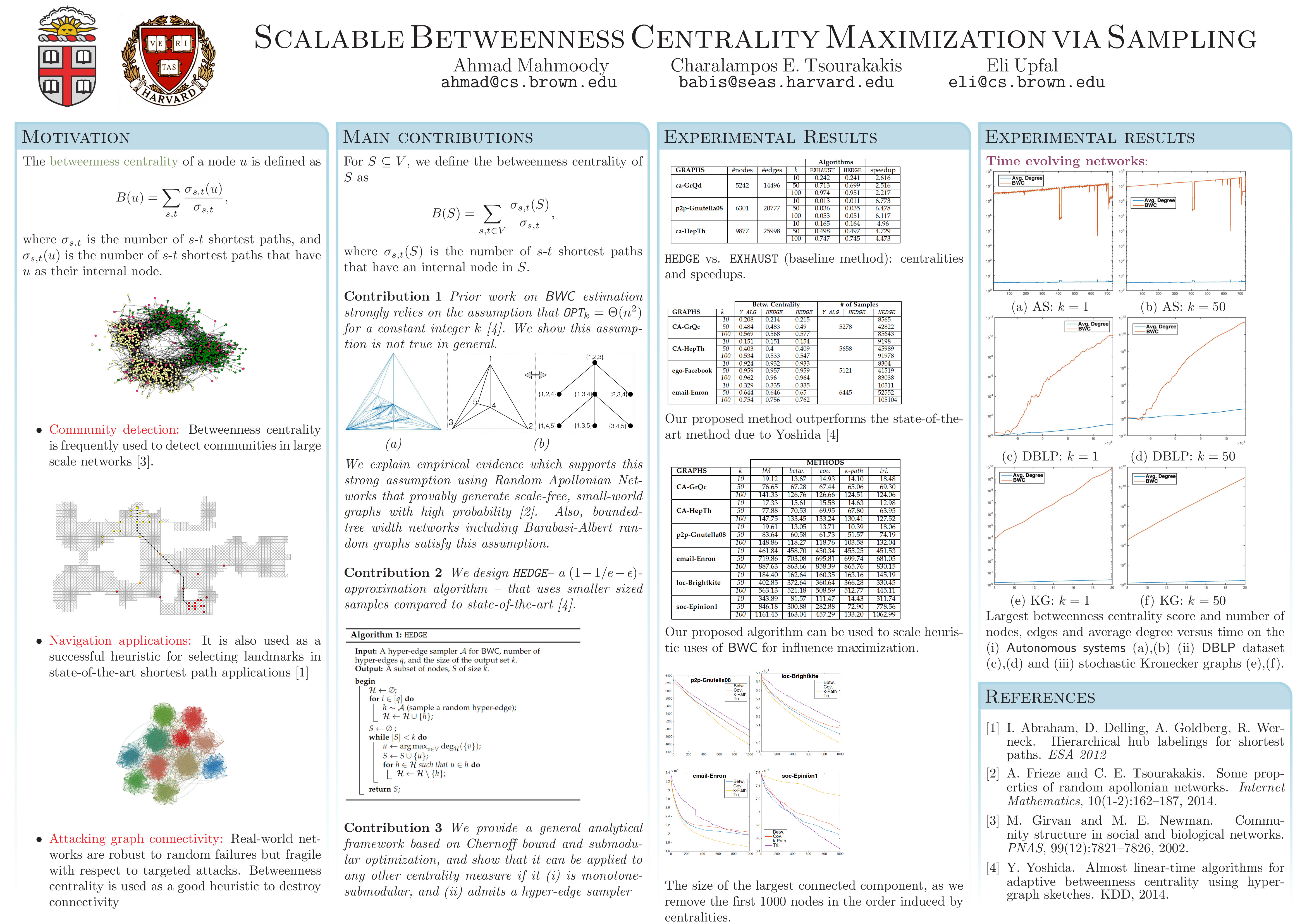
- A. Mahmoody, C. Tsourakakis, E. Upfal. Scalable Betweenness Centrality Maximization via Sampling, ACM KDD 2016.
- M. Ceccarello, A. Pietracaprina, G. Pucci, E. Upfal. A Practical Parallel Algorithm for Diameter Approximation of Massive Weighted Graphs, IPDPS 2016: 12-21.
- J. Augustine, W. K. Moses Jr., A. Redlich, E. Upfal. Balanced Allocation: Patience is not a Virtue, SODA 2016: 655-671.
- M. Riondato, E. M. Kornaropoulos. Fast Estimation of Betweenness Centrality through Sampling, Data Mining and Knowledge Discovery, 2016.
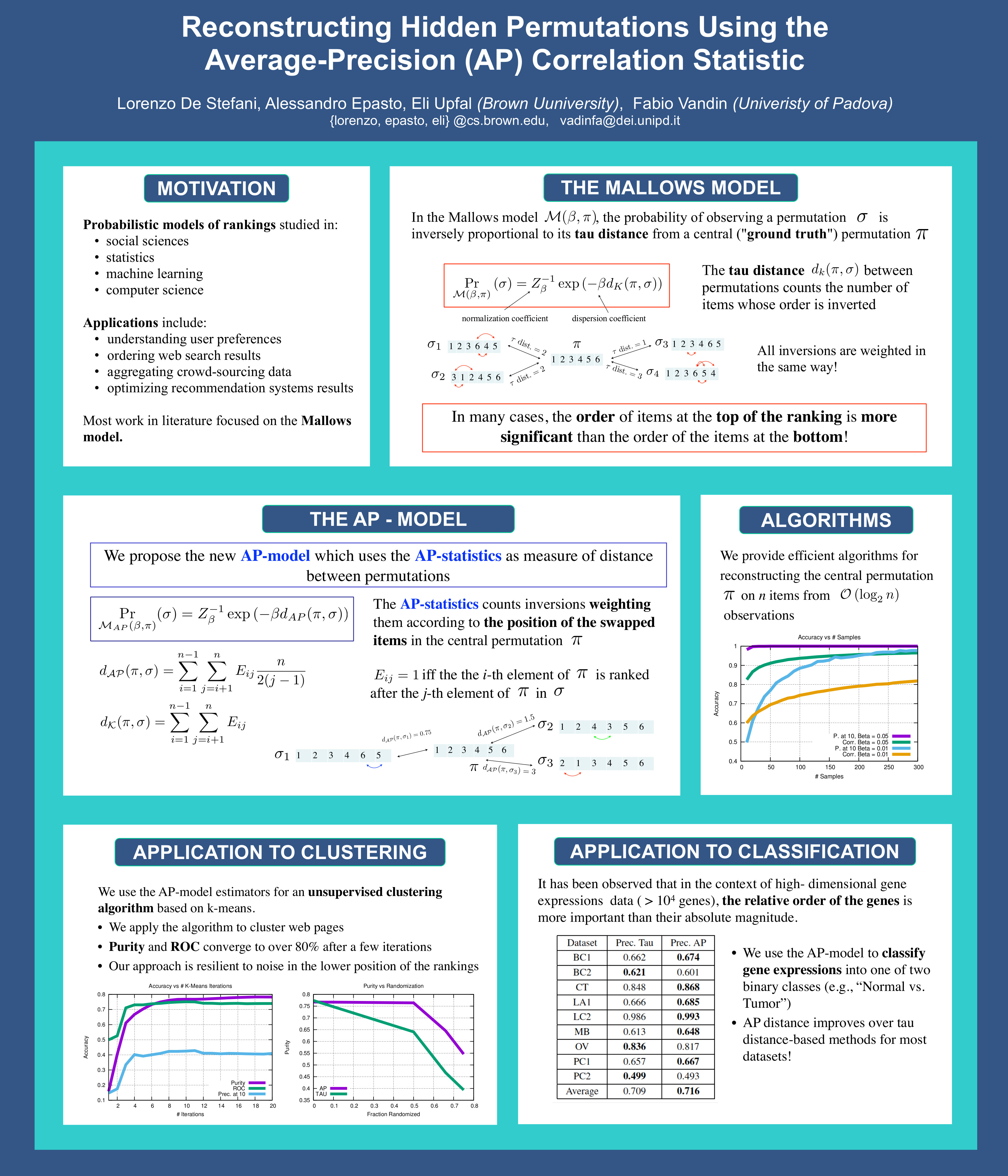
- L. De Stefani, A. Epasto, E. Upfal, F. Vandin. Reconstructing Hidden Permutations Using the Average-Precision (AP) Correlation Statistic, AAAI 2016.
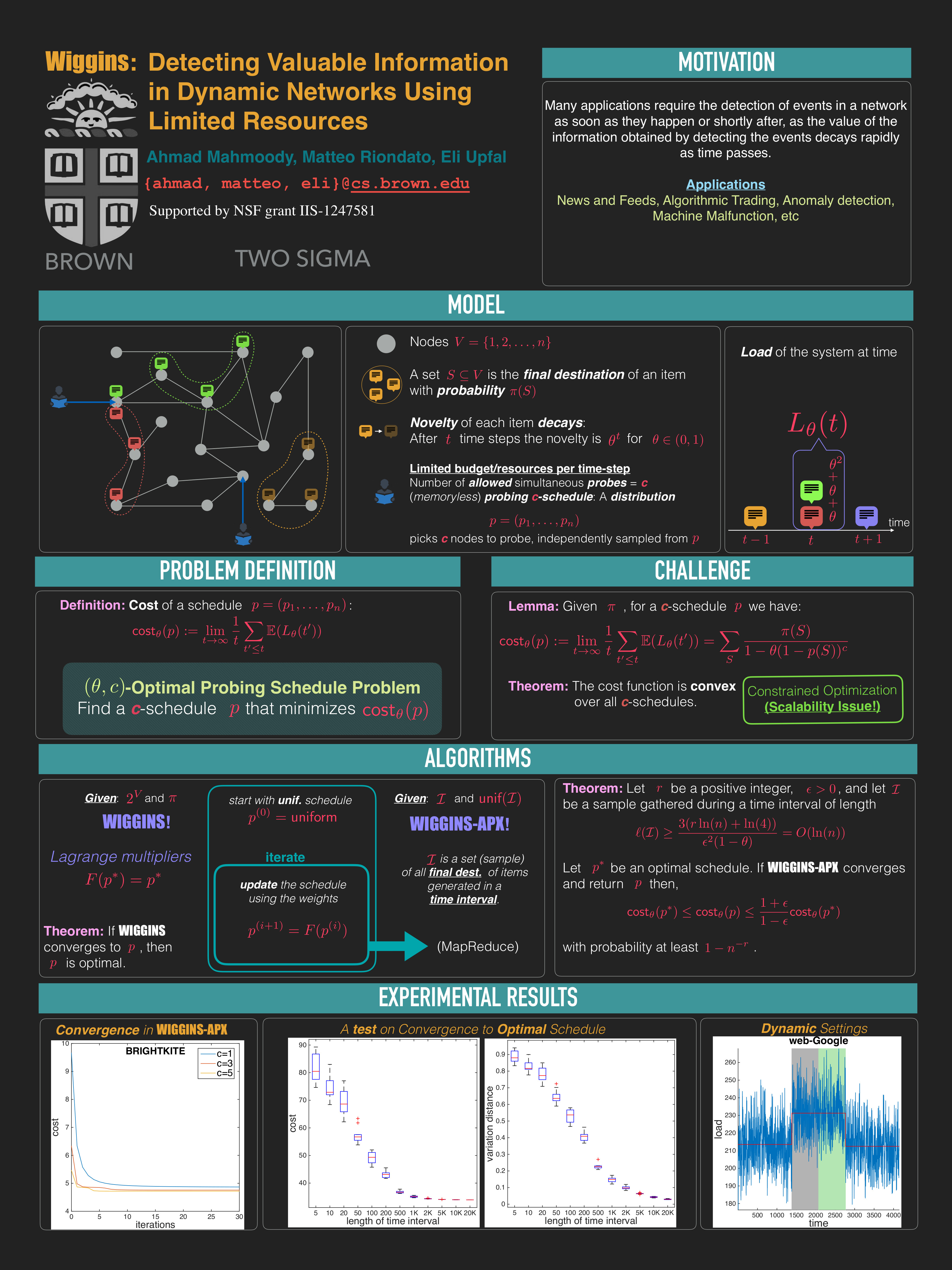
- A. Mahmoody, M. Riondato, E. Upfal. WIGGINS: Detecting Valuable Information in Dynamic Networks Using Limited Resources, ACM WSDM 2016.
- A. Das Sarma, A. Rahaman Molla, G. Pandurangan, E. Upfal. Fast distributed PageRank computation, Theor. Comput. Sci. 561: 113-121, 2015.
- J. Augustine, G. Pandurangan, P. Robinson, S. T. Roche, E. Upfal. Enabling Robust and Efficient Distributed Computation in Dynamic Peer-to-Peer Networks, FOCS 2015: 350-369.
- A. Mahmoody, E. M. Kornaropoulos, E. Upfal. Optimizing Static and Adaptive Probing Schedules for Rapid Event Detection, COCOA 2015.
- H. Wallace. Clustering of Musical Genres, Honor Thesis.
- M. Riondato, E. Upfal. VC-Dimension and Rademacher Averages: From Statistical Learning Theory to Sampling Algorithms — A Tutorial, ACM KDD 2015. Tutorial website.
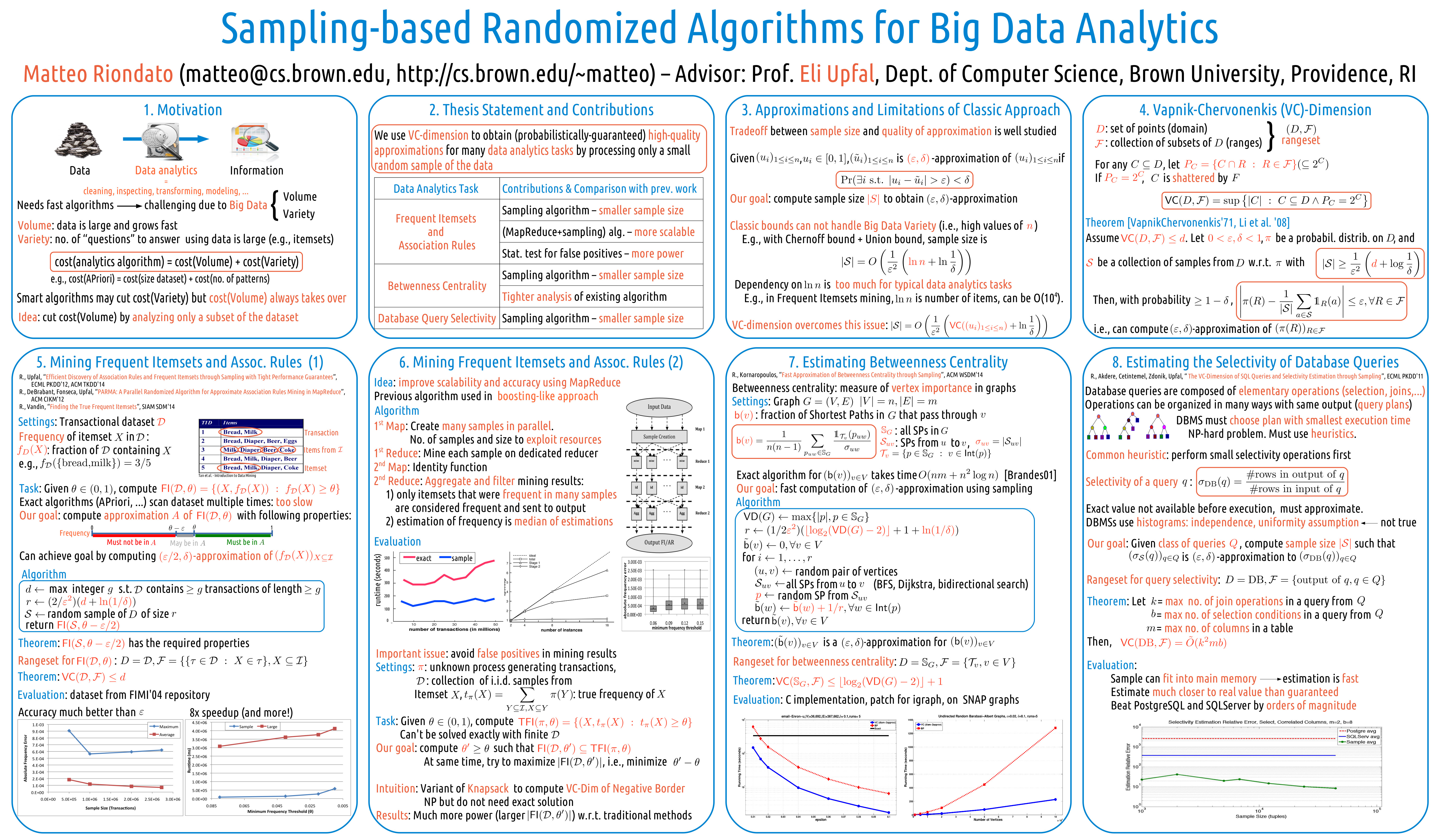
- M. Riondato, E. Upfal. Mining Frequent Itemsets through Progressive Sampling with Rademacher Averages, ACM KDD 2015.
- A. Epasto, S. Lattanzi, M. Sozio.Efficient Densest Subgraph Computation in Evolving Graphs, WWW 2015
- M. Ceccarello, A. Pietracaprina, G. Pucci, E. Upfal. Space and Time Efficient Parallel Graph Decomposition, Clustering, and Diameter Approximation , ACM SPAA 2015
- F. Vandin, A. Papoutsaki, B.J. Raphael, E. Upfal. Accurate Computation of Survival Statistics in Genome-Wide Studies, PLoS Computational Biology, 2015
- F. Vandin, B.J. Raphael, E. Upfal. On the Sample Complexity of Cancer Pathways Identification, RECOMB 2015
- J. Augustine, G. Pandurangan, P. Robinson, E. Upfal. Distributed agreement in dynamic peer-to-peer networks, Journal of Computer and System Sciences, 2015
- M. Riondato, D. García-Soriano, F. Bonchi. Graph Summarization with Quality Guarantees, IEEE ICDM 2014.
- M. Riondato, E. M. Kornaropoulos. Fast Estimation of Betweenness Centrality through Sampling, ACM WSDM 2014.
- M. Riondato, E. Upfal. Efficient Discovery of Association Rules and Frequent Itemsets through Sampling with Tight Performance Guarantees, ACM Transactions on Knowledge Discovery from Data, 2014.
- M. Riondato, F. Vandin. Finding the True Frequent Itemsets, SIAM SDM 2014.
Acknowledgements
The project is supported by NSF grant RI-1813444. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
The project is supported, in part, by DARPA/AFRL grant FA8750. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of DARPA/AFRL.
This project is supported, in part, by funding from Two Sigma Investments, LP. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflects the views of Two Sigma Investments, LP.